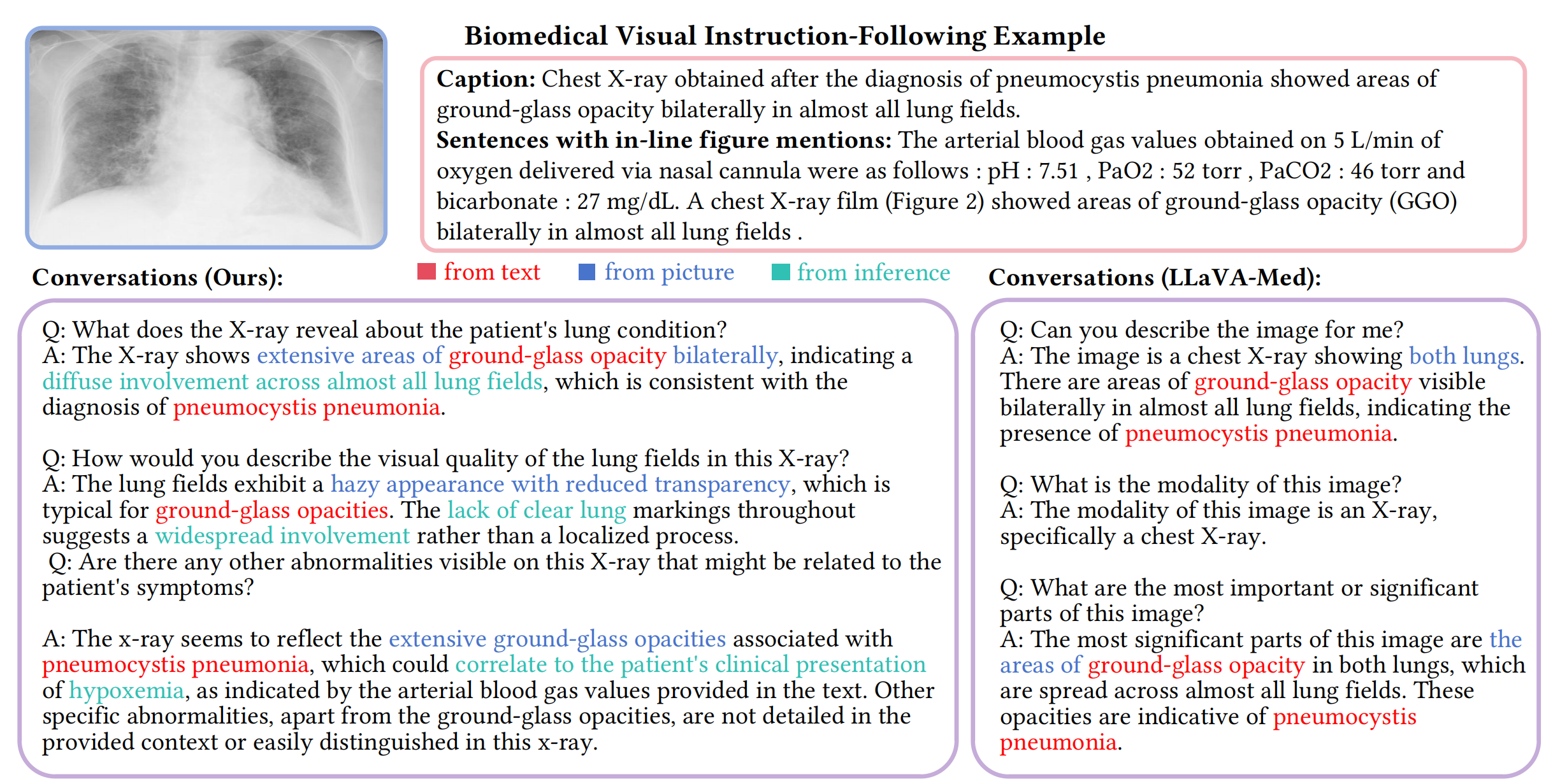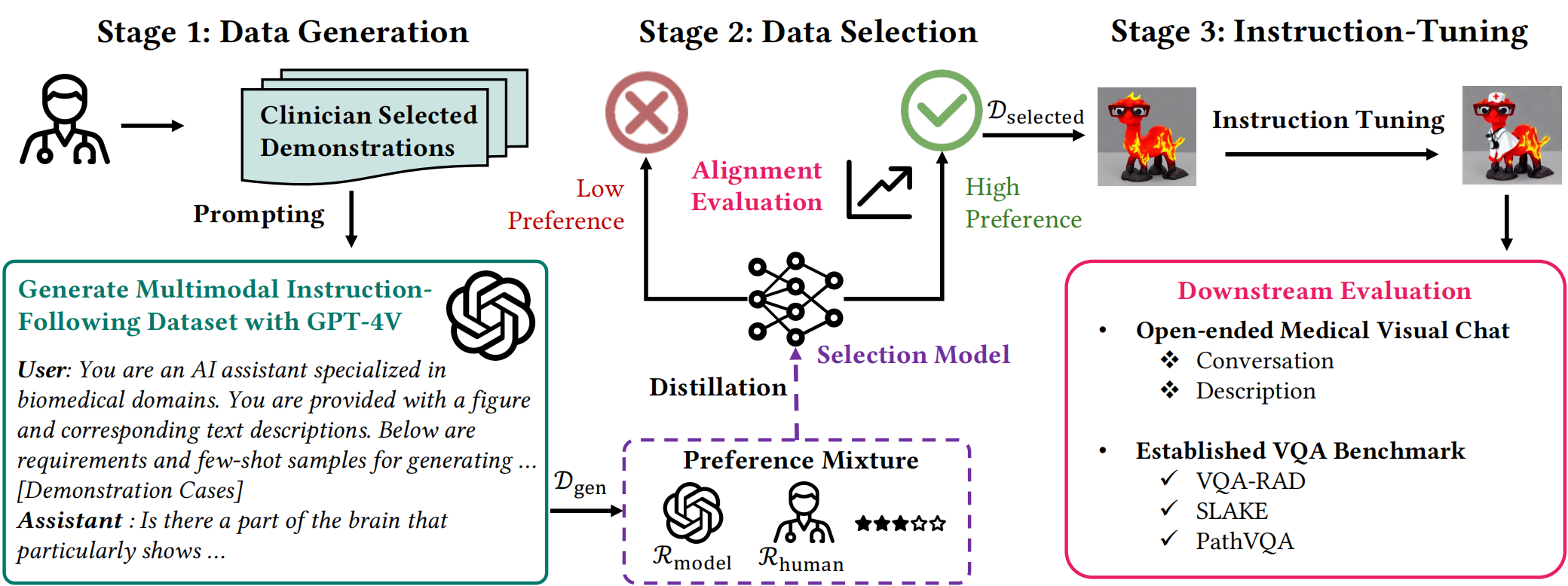
In this work, we propose a data-centric framework, Biomedical Visual Instruction Tuning with Clinician Preference Alignment (BiomedVITAl), that incorporates clinician preferences into both stages of generating and selecting instruction data for tuning biomedical multimodal foundation models. First, during the generation stage, we prompt the GPT-4V generator with a diverse set of clinician-selected demonstrations for preference-aligned data candidate generation. Then, during the selection phase, we train a separate selection model, which explicitly distills clinician and policy-guided model preferences into a rating function to select high-quality data for medical instruction tuning. Results show that the model tuned with the instruction-following data from our method demonstrates a significant improvement in open visual chat (18.5% relatively) and medical VQA (win rate up to 81.73%).
 Multimodal Medical Instrucion-Following Data
Multimodal Medical Instrucion-Following Data